Cusabio Human Recombinants
Recombinant Human Replication protein A 70kDa DNA-binding subunit (RPA1) | CSB-EP020088HU
- SKU:
- CSB-EP020088HU
- Availability:
- 3 - 7 Working Days
Description
Recombinant Human Replication protein A 70kDa DNA-binding subunit (RPA1) | CSB-EP020088HU | Cusabio
Alternative Name(s): Replication factor A protein 1 ;RF-A protein 1Single-stranded DNA-binding protein
Gene Names: RPA1
Research Areas: Epigenetics and Nuclear Signaling
Organism: Homo sapiens (Human)
AA Sequence: VGQLSEGAIAAIMQKGDTNIKPILQVINIRPITTGNSPPRYRLLMSDGLNTLSSFMLATQLNPLVEEEQLSSNCVCQIHRFIVNTLKDGRRVVILMELEVLKSAEAVGVKIGNPVPYNEGLGQPQVAPPAPAASPAASSRPQPQNGSSGMGSTVSKAYGASKTFGKAAGPSLSHTSGGTQSKVVPIASLTPYQSKWTICARVTNKSQIRTWSNSRGEGKLFSLELVDESGEIRATAFNEQVDKFFPLIEVNKVYYFSKGTLKIANKQFTAVKNDYEMTFNNETSVMPCEDDHHLPTVQFDFTGIDDLENKSKDSLVDIIGICKSYEDATKITVRSNNREVAKRNIYLMDTSGKVVTATLWGEDADKFDGSRQPVLAIKGARVSDFGGRSLSVLSSSTIIANPDIPEAYKLRGWFDAEGQALDGVSISDLKSGGVGGSNTNWKTLYEVKSENLGQGDKPDYFSSVATVVYLRKENCMYQACPTQDCNKKVIDQQNGLYRCEKCDTEFPNFKYRMILSVNIADFQENQWVTCFQESAEAILGQNAAYLGELKDKNEQAFEEVFQNANFRSFIFRVRVKVETYNDESRIKATVMDVKPVDYREYGRRLVMSIRRSALM
Source: E.coli
Tag Info: N-terminal 6xHis-SUMO-tagged
Expression Region: 2-616aa
Sequence Info: Full Length of Mature Protein
MW: 84 kDa
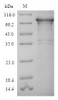
Purity: Greater than 90% as determined by SDS-PAGE.
Relevance: As part of the heterotrimeric replication protein A complex (RPA/RP-A), binds and stabilizes single-stranded DNA intermediates, that form during DNA replication or upon DNA stress. It prevents their reannealing and in parallel, recruits and activates different proteins and complexes involved in DNA metabolism. Thereby, it plays an essential role both in DNA replication and the cellular response to DNA damage . In the cellular response to DNA damage, the RPA complex controls DNA repair and DNA damage checkpoint activation. Through recruitment of ATRIP activates the ATR kinase a master regulator of the DNA damage response . It is required for the recruitment of the DNA double-strand break repair factors RAD51 and RAD52 to chromatin in response to DNA damage . Also recruits to sites of DNA damage proteins like XPA and XPG that are involved in nucleotide excision repair and is required for this mechanism of DNA repair . Plays also a role in base excision repair (BER) probably through interaction with UNG . Through RFWD3 may activate CHEK1 and play a role in replication checkpoint control. Also recruits SMARCAL1/HARP, which is involved in replication fork restart, to sites of DNA damage. May also play a role in telomere maintenance . As part of the alternative replication protein A complex, aRPA, binds single-stranded DNA and probably plays a role in DNA repair. Compared to the RPA2-containing, canonical RPA complex, may not support chromosomal DNA replication and cell cycle progression through S-phase. The aRPA may not promote efficient priming by DNA polymerase alpha but could support DNA synthesis by polymerase delta in presence of PCNA and replication factor C (RFC), the dual incision/excision reaction of nucleotide excision repair and RAD51-dependent strand exchange
Reference: Complete sequencing and characterization of 21,243 full-length human cDNAs.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Kimura K., Makita H., Sekine M., Obayashi M., Nishi T., Shibahara T., Tanaka T., Ishii S. , Yamamoto J., Saito K., Kawai Y., Isono Y., Nakamura Y., Nagahari K., Murakami K., Yasuda T., Iwayanagi T., Wagatsuma M., Shiratori A., Sudo H., Hosoiri T., Kaku Y., Kodaira H., Kondo H., Sugawara M., Takahashi M., Kanda K., Yokoi T., Furuya T., Kikkawa E., Omura Y., Abe K., Kamihara K., Katsuta N., Sato K., Tanikawa M., Yamazaki M., Ninomiya K., Ishibashi T., Yamashita H., Murakawa K., Fujimori K., Tanai H., Kimata M., Watanabe M., Hiraoka S., Chiba Y., Ishida S., Ono Y., Takiguchi S., Watanabe S., Yosida M., Hotuta T., Kusano J., Kanehori K., Takahashi-Fujii A., Hara H., Tanase T.-O., Nomura Y., Togiya S., Komai F., Hara R., Takeuchi K., Arita M., Imose N., Musashino K., Yuuki H., Oshima A., Sasaki N., Aotsuka S., Yoshikawa Y., Matsunawa H., Ichihara T., Shiohata N., Sano S., Moriya S., Momiyama H., Satoh N., Takami S., Terashima Y., Suzuki O., Nakagawa S., Senoh A., Mizoguchi H., Goto Y., Shimizu F., Wakebe H., Hishigaki H., Watanabe T., Sugiyama A., Takemoto M., Kawakami B., Yamazaki M., Watanabe K., Kumagai A., Itakura S., Fukuzumi Y., Fujimori Y., Komiyama M., Tashiro H., Tanigami A., Fujiwara T., Ono T., Yamada K., Fujii Y., Ozaki K., Hirao M., Ohmori Y., Kawabata A., Hikiji T., Kobatake N., Inagaki H., Ikema Y., Okamoto S., Okitani R., Kawakami T., Noguchi S., Itoh T., Shigeta K., Senba T., Matsumura K., Nakajima Y., Mizuno T., Morinaga M., Sasaki M., Togashi T., Oyama M., Hata H., Watanabe M., Komatsu T., Mizushima-Sugano J., Satoh T., Shirai Y., Takahashi Y., Nakagawa K., Okumura K., Nagase T., Nomura N., Kikuchi H., Masuho Y., Yamashita R., Nakai K., Yada T., Nakamura Y., Ohara O., Isogai T., Sugano S.Nat. Genet. 36:40-45(2004)
Storage: The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself. Generally, the shelf life of liquid form is 6 months at -20?/-80?. The shelf life of lyophilized form is 12 months at -20?/-80?.
Notes: Repeated freezing and thawing is not recommended. Store working aliquots at 4? for up to one week.
Function: As part of the heterotrimeric replication protein A complex (RPA/RP-A), binds and stabilizes single-stranded DNA intermediates, that form during DNA replication or upon DNA stress. It prevents their reannealing and in parallel, recruits and activates different proteins and complexes involved in DNA metabolism
Involvement in disease:
Subcellular Location: Nucleus, Nucleus, PML body
Protein Families: Replication factor A protein 1 family
Tissue Specificity:
Paythway: DNArepairpathway
Form: Liquid or Lyophilized powder
Buffer: If the delivery form is liquid, the default storage buffer is Tris/PBS-based buffer, 5%-50% glycerol. If the delivery form is lyophilized powder, the buffer before lyophilization is Tris/PBS-based buffer, 6% Trehalose, pH 8.0.
Reconstitution: We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20?/-80?. Our default final concentration of glycerol is 50%. Customers could use it as reference.
Uniprot ID: P27694
HGNC Database Link: HGNC
UniGene Database Link: UniGene
KEGG Database Link: KEGG
STRING Database Link: STRING
OMIM Database Link: OMIM