Pancreatic Cancer
Pancreatic Cancer
Pancreatic cancer is a disease in which malignant cells grow in the tissues of the pancreas.
The pancreas is an organ located horizontally behind the lower part of your stomach and in front of the spine that produces digestive juices and hormones to digest and absorb food nutrients.
A diagram of pancreas structure
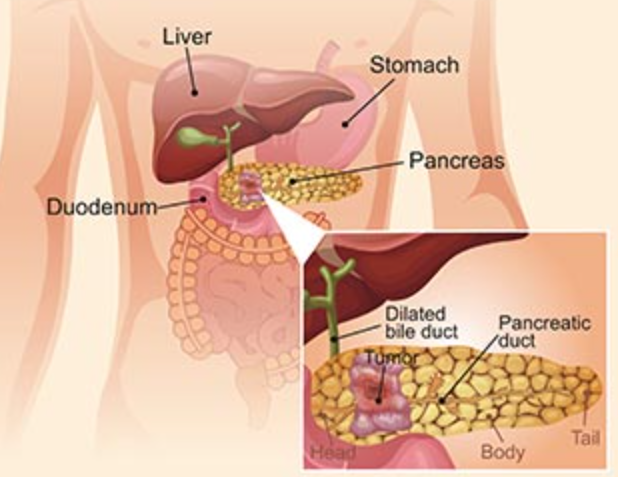
Figure 1. The pancreas structure
Pancreatic cancer occurs when pancreas cells develop abnormal DNA mutations in the pancreas that make them divide and grow a tumor.
Two types of growths can occur in the pancreas, including cancerous and noncancerous tumors.
- The most common type of cancer Pancreatic adenocarcinoma in the cells that line the ducts that carry digestive enzymes out of the pancreas (pancreatic ductal adenocarcinoma). Since the pancreas is located deep in the abdomen, pancreatic cancer is difficult to detect and diagnose. Tumors may cause symptoms including jaundice, pain, loss of appetite, pancreatitis, or unintended weight loss.
- Pancreatic neuroendocrine tumors (NETs) are a less common type and are discussed in Pancreatic Neuroendocrine Tumors.
The molecular pathology of pancreatic cancer is dominated by activating mutations in KRAS, which are present in >90% of tumors. Inactivating mutations of TP53, CDKN2A and SMAD4 occur in 50–80% of pancreatic cancers.
Other genes, including ARID1A and TGFBR2, are mutated in ~10% of tumors [2] [3] [4] [5]. In this article, we list part of targets involved in pancreatic cancer based on the information provided by NCG (web resource to analyze duplicability, orthology and network properties of cancer genes)
| ABCA7 | ABCC5 | ABLIM2 | ACSM1 | ACVR1B | ACVR2A | ADAMTS20 | ADGRB3 |
| ADGRD1 | ADGRL1 | AKAP11 | AKAP13 | ALMS1 | ARID1A | ARID2 | ASL |
| ATM | ATP10A | ATXN1 | BCLAF1 | BCORL1 | BRCA2 | CDKN2A | GNAS |
| KDM6A | KMT2C | KRAS | MACF1 | MAP2K4 | PBRM1 | PREX2 | RBM10 |
| RNF43 | SF3B1 | SMAD4 | SMARCA4 | TGFBR2 | TP53 |
Here, we display several key targets involved in molecular pathology of pancreatic cancer, including:
- CDKN2A (cyclin-dependent kinase inhibitor 2A), also known as p16-INK4a, MTS-1, or CDK4I, is the tumor suppressor, which regulates cell cycle progression by inhibiting cyclinD-CDK4 and cyclinD-CDK6 complexes responsible for initiating the G1/S phase transition. It is one of the most frequently altered genes in cancer, and the incidence of mutations in sporadic pancreatic cancer is impressive, with inactivation occurring in 98% of cases [6] [7].
- SMAD4 (SMAD Family Member 4), a tumor suppressor protein, translocates to the nucleus as heterotrimeric SMAD2/SMAD3-SMAD4 complex after TGFβ family receptors activation, where it activates the expression of genes and causes growth inhibition [8]. In pancreatic ductal adenocarcinoma, four driver genes, namely, KRAS, TP53, SMAD4, and CDKN2A, have been reported as representative cancer-related genes [9]. Moreover, mutations of SMAD4 occur in around 50% of pancreatic cancers, mostly leading to the loss of protein activated.
- ARID1A (AT-rich interactive domain containing protein 1A), a part of the SWI/SNF complex, is a multi-subunit complex that remodels chromatin by shifting, inserting or evicting nucleosomes in an ATP-dependent manner. ARID1A is commonly mutated in pancreatic ductal adenocarcinoma, and emerging study has demonstrated that ARID1A suppresses pancreatic neoplasia in a compartment-specific manner. Moreover, in duct cells, this process appears to be associated with MYC-facilitated protein synthesis [10].
- KMT2C (Lysine N-methyltransferase 2C), also known as myeloid/lymphoid or mixed-lineage leukemia protein 3, is a member of KMT2 family of histone lysine methyltransferases which consists of KMT2A, KMT2B, KMT2C, KMT2D, KMT2E, KMT2F, KMT2G, and KMT2H [11]. Among of them, KMT2C and KMT2D are mono-methyltransferases in the COMPASS complex, both of which are target H3K4 for epigenetic regulation [12]. Several studies have observed KMT2C (missense) mutations and KMT2D (missense) mutations in patients with pancreatic cancer. Furthermore, nonsense/missense mutations in KMT2C/D correlate with a better prognosis of patients with pancreatic cancer, as reported by Sausen et al [13].
- RNF43 (ring finger 43), as a transmembrane E3 ubiquitin ligase, downregulates the Wnt/β-catenin pathway through ubiquitinating the Wnt receptor frizzled [14]. Mutations of RNF43 have been identified in various tumors, including cystic pancreatic tumors.
References
[1] Alexandra Moore and Timothy Donahue. Pancreatic Cancer [J]. JAMA. 2019, 322 (14): 1426.
[2] Wang, L. et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency [J]. Genome Res. 2012, 22, 208–219.
[3] Waddell, N. et al. Whole genomes redefine the mutational landscape of pancreatic cancer [J]. Nature. 2015, 518, 495–501.
[4] Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer [J]. Nature. 2016, 531, 47–52.
[5] Jorg Kleeff, Murray Korc, Minoti Apte et al. Pancreatic cancer [J]. Nat Rev Dis Primers. 2016, 21 (2):16022.
[6] Schutte, M.; Hruban, R.H.; Geradts, J. et al. Abrogation of the RB/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas [J]. Cancer Res. 1997, 57, 3126–3130.
[7] Jonas Cicenas, Kotryna Kvederaviciute, Ingrida Meskinyte et al. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer [J]. Cancers. 2017, 9 (12): 42–.
[8] Cao, D.; Ashfaq, R.; Goggins, M.G. et al. Differential expression of multiple genes in association with MADH4/DPC4/SMAD4 inactivation in pancreatic cancer [J]. Int. J. Clin. Exp. Pathol. 2008, 1, 510–517.
[9] Takahiro Yokose, Minoru Kitago, Sachiko Matsuda et al. Combination of KRAS and SMAD4 mutations in formalin-fixed paraffin-embedded tissues as a biomarker for pancreatic cancer [J]. Cancer Sci. 2020, 111(6):2174-2182.
[10] Sam C Wang, Ibrahim Nassour, Shu Xiao et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation [J]. Gut. 2019, 68:1259-1270.
[11] Allis CD, Berger SL, Cote J et al. New nomenclature for chromatin-modifying enzymes [J]. Cell. 2007, 131:633–6.
[12] Rokutan, H.; Hosoda, F.; Hama, N. et al. Comprehensive mutation profiling of mucinous gastric carcinoma [J]. J. Pathol. 2016, 240, 137–148.
[13] Sausen, M.; Phallen, J.; Adleff, V. et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients [J]. Nat. Commun. 2015, 6, 7686.
[14] Xiao Yan Chang, Yan Wu, Ying Jiang et al. RNF43 Mutations in IPMN Cases: A Potential Prognostic Factor [J]. Gastroenterology Research and Practice. 2020, 1457452.
Recent Posts
-
Cusabio custom services
Cusabio offers custom services through Gentaur in the field of life sciences, specifically in the pr …3rd Feb 2024 -
Pancreatic Cancer
Pancreatic CancerPancreatic cancer is a disease in which malignant cells grow in the tissues of the …1st Nov 2021

